Example: Bioreactor Model
Here we show how bioreaction systems can be modeled in REX. A batch fermentation of glucose by Aspergillus niger to produce gluconic acid is studied based on the work of Fatmawati et al. [1].
The batch model proposed in the paper consists of the following equations:
Cell Growth
where [Cell] represents the mass concentration (g/l) of cells, t refers to time (h), μm is the maximum specific growth rate (/h) and [Cell]m is the maximum cell concentration (g/l).
Product Formation
where [Product] is the product concentration of gluconic acid (g/l), ⍺ is the cell growth associated product formation constant and β is the product formation constant (/h) irrespective of cell growth. Since this is a pure batch reactor (no feeds or outflows), the cell growth rate and the time derivative of the reactor cell concentration are identical.
Glucose Consumption
This simplified model considers that the glucose substrate is consumed for cell mass growth and to sustain the existing cells (maintenance term). Direct consumption of Glucose for product formation is not considered:
where [Glucose] is the substrate concentration (g/l), YCell/Gluc is the yield of glucose to cell mass, and ms is the cell maintenance coefficient (/h).
Using measurements for [Cell], [Product] and [Glucose] along time, Fatmawati et al. [1] estimated the parameters in the equations above: μm, [Cell]m, ⍺, β, YCell/Gluc and ms.
Modeling the system in REX: First Considerations
The batch model proposed in the paper consists of the following equations:
Cell Growth
| [1] |
where [Cell] represents the mass concentration (g/l) of cells, t refers to time (h), μm is the maximum specific growth rate (/h) and [Cell]m is the maximum cell concentration (g/l).
Product Formation
| [2] |
where [Product] is the product concentration of gluconic acid (g/l), ⍺ is the cell growth associated product formation constant and β is the product formation constant (/h) irrespective of cell growth. Since this is a pure batch reactor (no feeds or outflows), the cell growth rate and the time derivative of the reactor cell concentration are identical.
Glucose Consumption
This simplified model considers that the glucose substrate is consumed for cell mass growth and to sustain the existing cells (maintenance term). Direct consumption of Glucose for product formation is not considered:
| [3] |
Using measurements for [Cell], [Product] and [Glucose] along time, Fatmawati et al. [1] estimated the parameters in the equations above: μm, [Cell]m, ⍺, β, YCell/Gluc and ms.
As observed in the equations above, the compounds are all treated in mass units. Thus, Concentrations = Mass Density should be selected in the Units Configurations node in order to change the project from the default molar basis to mass basis:
In the screen above, all the other relevant units have been selected to be consistent with Fatmawati et al. [1].
Another issue is that the model equations posed in [1] is for concentration derivatives. However, the REX internal model works with derivatives for total mass of compounds, so for batch we have the general mass balance:
where XiTM represent total mass of compound i, while V is the reactor volume, Rater is the rate for reaction r, and vi,r is the stoichiometric coefficient for compound i in reaction r.
Nevertheless, for constant volume we can convert to concentration derivative as shown below:
After canceling V, we get the mass balance expressed in term of concentration derivative, same as in the proposed model equations :
Consequently, we can set unit volume in equation 4 above and model the system in REX. REX will internally treat the numbers as mass, but the model is now the same as the original formulation in [1] because of the unit volume assumption.
Defining Bioreactor Reactions in REX
|
|
|---|
In the screen above, all the other relevant units have been selected to be consistent with Fatmawati et al. [1].
Another issue is that the model equations posed in [1] is for concentration derivatives. However, the REX internal model works with derivatives for total mass of compounds, so for batch we have the general mass balance:
| [4] |
where XiTM represent total mass of compound i, while V is the reactor volume, Rater is the rate for reaction r, and vi,r is the stoichiometric coefficient for compound i in reaction r.
Nevertheless, for constant volume we can convert to concentration derivative as shown below:
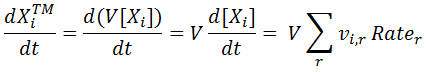 |
[5] |
After canceling V, we get the mass balance expressed in term of concentration derivative, same as in the proposed model equations :
| [6] |
Consequently, we can set unit volume in equation 4 above and model the system in REX. REX will internally treat the numbers as mass, but the model is now the same as the original formulation in [1] because of the unit volume assumption.
You may follow this section by looking at the Bioreactor-1.rex file that is located in the Optience Corporation\Rex Suite\Examples folder.
One issue you may have realized is that equations [2] and [3] are related to equation [1]. You can see that glucose consumption has a term related to cell growth, and are they tied by the yield coefficient YCell/Gluc. We start by defining the reactions in REX independently and then enforce their dependency through parameter relationships. This approach is simple in design - we first present his approach and later show an alternative formulation that is more compact and preferable.
When analyzing equation [1] for cell growth, we see that it can be modeled in REX by a single reaction with no reactant and only cell as product. When a reaction has only products or reactants, REX marks these reaction equations with a formation or consumption suffix respectively. So in this case Reaction Equation is named Cell Formation:
In the Kinetics node we include forward and reverse directions in order to model both terms from equation [1] when expanded to:
Then in Parameters node we define the reaction orders, that are order 1 for Cell in the forward direction and order 2 for Cell in the reverse direction.
With this setup, the preexponential of the forward direction will be same as μm, while the preexponential for the reverse direction will be equivalent to μm / [Cellm]
As for Product formation [2], there are two terms that we should deal with in two different REX reactions.
The first term of equation [2] is tied to cell growth: that term is exactly same as equation [1] times ⍺. Thus, for that term we define a reaction Rprod-alpha in REX where only Product is in its stoichiometry:
Then we do same as in the Cell formation reaction R1: both directions are included and orders for Cell are defined as one and two for the forward and reverse directions respectively.
From this formulation, the preexponential of the forward direction will be same as ⍺ μm, while the preexponential for the reverse direction will be equivalent to ⍺ μm / [Cellm]
As for the second term of equation [2], we model as another reaction Rprod-beta, with same stoichiometry than Rprod-alpha, but only forward direction included. That direction has order one with respect to Cells.
Finally, the glucose consumption reaction is modeled quite similar as the product formation. Each term is dealt with by a different REX reaction; the main difference is that both reactions consider glucose as reactant to account for the negative sign on [3]:
Reaction RGc is used for the first term of equation [3] that represent the consumption of substrate to produce cell mass. Thus both directions are included, using orders for cell of one and two for the forward and reverse directions respectively.
The preexponential of the forward direction will be same as μm / YCell/Gluc, while the preexponential for the reverse direction will be equivalent to μm / [Cellm] / YCell/Gluc.
The second term of equation [3] is the glucose consumption for cell maintenance, and it is modelled in REX by reaction RGm. That reaction has only forward direction included, with order one with respect to Cell.
We must now enforce some parameter relationships for the preexponentials that are related to the Cell growth.
As discussed, both the Glucose consumption and Product formation has one term that is proportional to the Cell growth. We must ensure that the preexponential ratio between forward and reverse direction is same for the reactions that are related to the cell growth. The Parameter Relationships node view is shown below:
Alternative REX formulation
One issue you may have realized is that equations [2] and [3] are related to equation [1]. You can see that glucose consumption has a term related to cell growth, and are they tied by the yield coefficient YCell/Gluc. We start by defining the reactions in REX independently and then enforce their dependency through parameter relationships. This approach is simple in design - we first present his approach and later show an alternative formulation that is more compact and preferable.
When analyzing equation [1] for cell growth, we see that it can be modeled in REX by a single reaction with no reactant and only cell as product. When a reaction has only products or reactants, REX marks these reaction equations with a formation or consumption suffix respectively. So in this case Reaction Equation is named Cell Formation:
|
|
|---|
In the Kinetics node we include forward and reverse directions in order to model both terms from equation [1] when expanded to:
| [7] |
Then in Parameters node we define the reaction orders, that are order 1 for Cell in the forward direction and order 2 for Cell in the reverse direction.
With this setup, the preexponential of the forward direction will be same as μm, while the preexponential for the reverse direction will be equivalent to μm / [Cellm]
As for Product formation [2], there are two terms that we should deal with in two different REX reactions.
The first term of equation [2] is tied to cell growth: that term is exactly same as equation [1] times ⍺. Thus, for that term we define a reaction Rprod-alpha in REX where only Product is in its stoichiometry:
|
|
|---|
Then we do same as in the Cell formation reaction R1: both directions are included and orders for Cell are defined as one and two for the forward and reverse directions respectively.
From this formulation, the preexponential of the forward direction will be same as ⍺ μm, while the preexponential for the reverse direction will be equivalent to ⍺ μm / [Cellm]
As for the second term of equation [2], we model as another reaction Rprod-beta, with same stoichiometry than Rprod-alpha, but only forward direction included. That direction has order one with respect to Cells.
Finally, the glucose consumption reaction is modeled quite similar as the product formation. Each term is dealt with by a different REX reaction; the main difference is that both reactions consider glucose as reactant to account for the negative sign on [3]:
|
|
|---|
Reaction RGc is used for the first term of equation [3] that represent the consumption of substrate to produce cell mass. Thus both directions are included, using orders for cell of one and two for the forward and reverse directions respectively.
The preexponential of the forward direction will be same as μm / YCell/Gluc, while the preexponential for the reverse direction will be equivalent to μm / [Cellm] / YCell/Gluc.
The second term of equation [3] is the glucose consumption for cell maintenance, and it is modelled in REX by reaction RGm. That reaction has only forward direction included, with order one with respect to Cell.
We must now enforce some parameter relationships for the preexponentials that are related to the Cell growth.
As discussed, both the Glucose consumption and Product formation has one term that is proportional to the Cell growth. We must ensure that the preexponential ratio between forward and reverse direction is same for the reactions that are related to the cell growth. The Parameter Relationships node view is shown below:
|
|
|---|
There is a more compact way to define this model in REX. You can import the Bioreactor-2.rex file located in folder Optience Corporation\Rex Suite\Examples to follow this section.
As mentioned in the previous section, both Product formation [2] and Glucose consumption [3] have a term that is related to the Cell growth reaction [1].
Thus, from the first term of [2] we can state that for each gram of cell created, Product increases by ⍺ grams. Similarly for the first term of [2]: for each gram of cell increase, 1 / YCell/Gluc grams of glucose is consumed.
Thus, we can merge reactions R1, Rprod-alpha and RGc from the previous section into a single reaction whose stoichiometry would be:
This new reaction has the advantage that we no longer need to enforce parameter relationships between the R1, Rprod-alpha and RGc used in the earlier formulation.
However, the stoichiometic coefficients for [8] are unknown, because both ⍺ and YCell/Gluc are parameters that must be estimated. We now take advantage of the stoichiometry estimation feature in REX by enabling it in the Estimation node:
Once the Estimation of Stoichiometry is enabled, a Stoichiometry tab is shown in the Estimation → Parameters node. Here, we open the stoichiometric bounds for Glucose and Product for the merged reaction. It is important to fix at least one of the stoichiometric coefficients to 1 to avoid non-unique parameters. In this case, we set the Cell to have unit coefficient. One advantage with this formulation is that once the Glucose coefficient is known, we can easily see how much Glucose is 'wasted' in the bioreactor. For example, in the above figure, about 11.15 grams (12.1508 - 1 - 0.00109757) of glucose are used for purposes other than cell growth and product formation:
As mentioned in the previous section, both Product formation [2] and Glucose consumption [3] have a term that is related to the Cell growth reaction [1].
Thus, from the first term of [2] we can state that for each gram of cell created, Product increases by ⍺ grams. Similarly for the first term of [2]: for each gram of cell increase, 1 / YCell/Gluc grams of glucose is consumed.
Thus, we can merge reactions R1, Rprod-alpha and RGc from the previous section into a single reaction whose stoichiometry would be:
| [8] |
This new reaction has the advantage that we no longer need to enforce parameter relationships between the R1, Rprod-alpha and RGc used in the earlier formulation.
However, the stoichiometic coefficients for [8] are unknown, because both ⍺ and YCell/Gluc are parameters that must be estimated. We now take advantage of the stoichiometry estimation feature in REX by enabling it in the Estimation node:
Once the Estimation of Stoichiometry is enabled, a Stoichiometry tab is shown in the Estimation → Parameters node. Here, we open the stoichiometric bounds for Glucose and Product for the merged reaction. It is important to fix at least one of the stoichiometric coefficients to 1 to avoid non-unique parameters. In this case, we set the Cell to have unit coefficient. One advantage with this formulation is that once the Glucose coefficient is known, we can easily see how much Glucose is 'wasted' in the bioreactor. For example, in the above figure, about 11.15 grams (12.1508 - 1 - 0.00109757) of glucose are used for purposes other than cell growth and product formation:
Parameter Estimation: Results
Both REX formulations have the same experimental data loaded in Experiments, that correspond to an initial condition of 150 g/l of Glucose and 10.34 g/l of Cells. Hybrid weights values were generated in REX to reconcile Cell, Product and Glucose measurements. After running both REX projects, you can verify that the fit to data is the same by comparing the weighted LSQ errors and the profile values for the compounds. Below we show the estimated parameter values from each REX implementation:
In both projects, μm and [Cell]m has the same value, while maintenance rate goes to zero. The YCell/Gluc yield from Bioreactor-2.rex file is the inverse of stoichiometric coefficient for glucose in Cellgrowth reaction, and its estimated value is 0.0823. That is the same value as we get for Bioreactor-1.rex obtained from the ratio between R1 and RGc forward preexponentials.
Predictions for Product are identical, as related parameters ⍺ and β are same in both REX implementations.
The parameter values obtained are somewhat different from Fatmawati et al. [1]. The reasons for this difference could be:
1. Fatmawati, A. and Agustriyanto. R., Modeling of Continuous Gluconic Acid Production by Fermentation, Science Journal, Ubon Ratchathani University 2010 1 (1), 82-89.
Top of Topic
In both projects, μm and [Cell]m has the same value, while maintenance rate goes to zero. The YCell/Gluc yield from Bioreactor-2.rex file is the inverse of stoichiometric coefficient for glucose in Cellgrowth reaction, and its estimated value is 0.0823. That is the same value as we get for Bioreactor-1.rex obtained from the ratio between R1 and RGc forward preexponentials.
Predictions for Product are identical, as related parameters ⍺ and β are same in both REX implementations.
The parameter values obtained are somewhat different from Fatmawati et al. [1]. The reasons for this difference could be:
- Weighting strategy: We used Hybrid weights for the measured values of all species. We do not know the weighting method used in [1].
- Maintenance coefficient ms : We allow this to be freely optimized and the solution goes to zero. The authors obtained a nonzero value, but they get a zero estimated value when reconciling data for other experiments where initial glucose concentration are 200 or 250 g/l.
- Initial Concentrations: We keep them fixed to experimental values. On the other hand, the authors appear to estimate them for Glucose and Cell mass. This can increase parameter non-uniqueness.
1. Fatmawati, A. and Agustriyanto. R., Modeling of Continuous Gluconic Acid Production by Fermentation, Science Journal, Ubon Ratchathani University 2010 1 (1), 82-89.
Go back to: